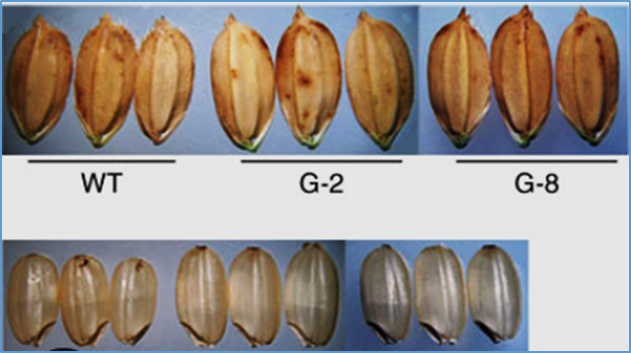
The regulatory role of ZmSTOMAGEN1/2 in maize stomatal development is elucidated via gene editing and metabolic profiling
Juan-Juan Xu, Qing-Yun Chen, Li-Fang Qin, Yuan Liu, You-Zhi Li, Xian-Wei Fan
PLOS One; July 14, 2025; https://doi.org/10.1371/journal.pone.0328433
Abstract
Stomatal development is mediated by EPIDERMAL PATTERNING FACTORs (EPFs), a family of secreted peptides including STOMAGEN/EPFL9 in Arabidopsis. To clarify the functional role of STOMAGEN orthologues in maize (Zea mays), we generated a double knockout mutant of ZmSTOMAGEN1 and ZmSTOMAGEN2 using CRISPR/Cas9 system. Comprehensive phenotypic analysis revealed that the zmstomagen1/2 mutant exhibited severe stomatal development defects, including complete absence of stomata between epidermal cells in stomatal lineage files and abnormal stomatal complexes with small lobed cells. These aberrant cells likely arose from failed asymmetric divisions of guard mother cells, ultimately preventing the formation of functional stomatal complexes. A double knockout of ZmSTOMAGEN1/2 reduced the expression of SPEECHLESS1 (SPCH1), MUTE, SCREAM2 (SCRM2), and STOMATAL DENSITY AND DISTRIBUTION1 (SDD1), impairing stomatal initiation and cell fate transition in early stomatal lineage cells. The mutant displayed a lower stomatal density and index, leading to reduced net photosynthetic rate, transpiration rate, and stomatal conductance but increased water-use efficiency (WUE). Compared to the wild-type plants (HiII-A × HiII-B), the zmstomagen1/2 mutant exhibited significant alterations in phytohormone homeostasis. These included brassinosteroid metabolite imbalance (increased typhasterol, decreased castasterone) and differential gibberellin regulation (elevated GA4, reduced GA1). These hormonal perturbations suggest that impaired stomatal morphogenesis in zmstomagen1/2 mutants result from disrupted crosstalk between multiple hormonals signaling networks. Our findings reveal a crucial role for ZmSTOMAGEN1/2 in regulating cell fate decisions within the stomatal lineage and provide a potential strategy for enhancing WUE in maize by manipulating ZmSTOMAGEN1/2 expression.
See https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0328433
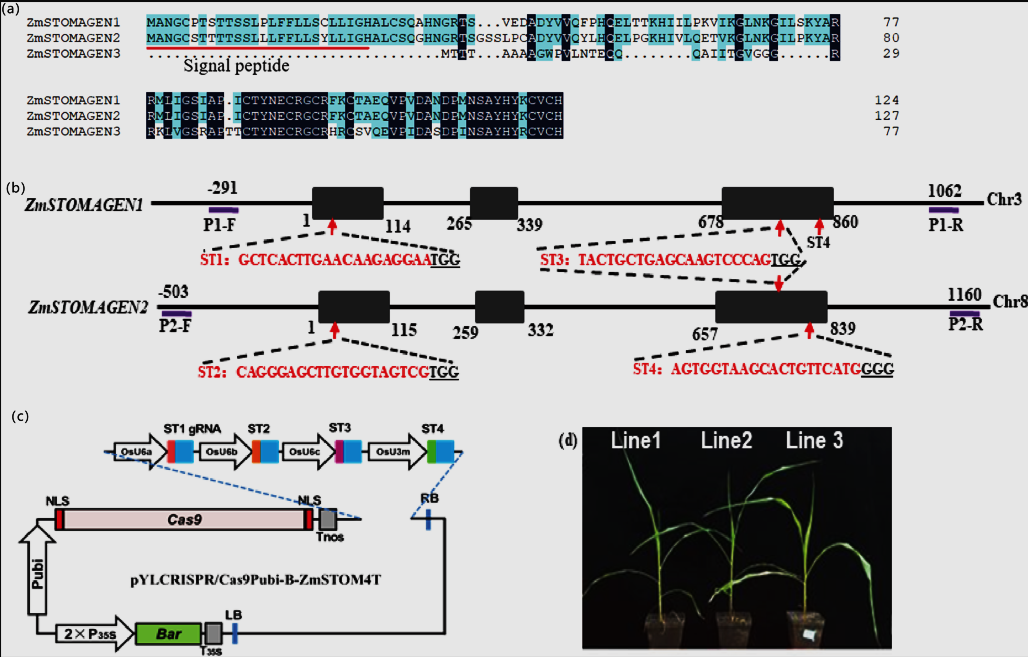
Figure 1
Schematic diagram of CRISPR/Cas9 system construction and transformation.
(a) A comparative analysis of the protein sequence of ZmSTOMAGEN1, ZmSTOMAGEN2 and ZmSTOMAGEN3. ZmSTOMAGEN1 and ZmSTOMAGEN2 share 86.99% sequence similarity, especially in the conserved functional domain (amino acid positions 79-123 of ZmSTOMAGEN1). However, these three ZmSTOMAGEN homologs are located on distinct chromosomes. (b) Schematic illustration of target genes and relative positions of gRNA binding sites in ZmSTOMAGEN1 and ZmSTOMAGEN2, respectively. Black box represents exons of ZmSTOMAGEN1 gene and ZmSTOMAGEN2 gene, respectively. Red arrow presents target site and the red sequence means the target sequence. Underline shows the PAM site for the gRNAs. Two pairs of primers (P1-F/P1-R) was designed to identify the sequence of ZmSTOMAGNEN1; P2-F/P2-R was used to identify the sequence of ZmSTOMAGEN2. The number represents nucleotide position. (c) Illustration of cloning of four sgRNA expression cassettes into CRISPR/Cas9 binary vectors by single Golden Gate ligation. (d) Transgenic plants (T0) were generated through the progeny of Hi-II maize immature embryos mediated by Agrobacterium tumefaciens.
https://doi.org/10.1371/journal.pone.0328433.g001
Số lần xem: 9
-
{mota}
-
{mota}
-
{mota}
-
{mota}
-
Biochar in the circular bionutrient economy
{mota} -
{mota}
-
Progress in Transcriptomics and Metabolomics in Plant Responses to Abiotic Stresses
{mota} -
{mota}
-
{mota}
-
Fine mapping and prediction of a candidate gene for wrinkled rind in melon (Cucumis melo L.)
{mota}